Episode Transcript
Interviewer: We're talking about Bam Iobio, an easy to use web-based app that analyzes genomic sequence data in seconds.
Announcer: Examining the latest research and telling you about the latest breakthroughs, the Science and Research Show is on The Scope.
Interviewer: I'm talking with Dr. Gabor Marth, professor of human genetics at the University of Utah. Dr. Marth, your app, called Bam Iobio, was just published in "Nature Methods." Why are you so excited about it?
Dr. Gabor Marth: Super excited about Bam.iobio because this is the first app that shows off the capabilities of this new interactive genomic ecosystem that we are building here at the University of Utah.
Interviewer: Tell me, what is Bam Iobio? What does it do?
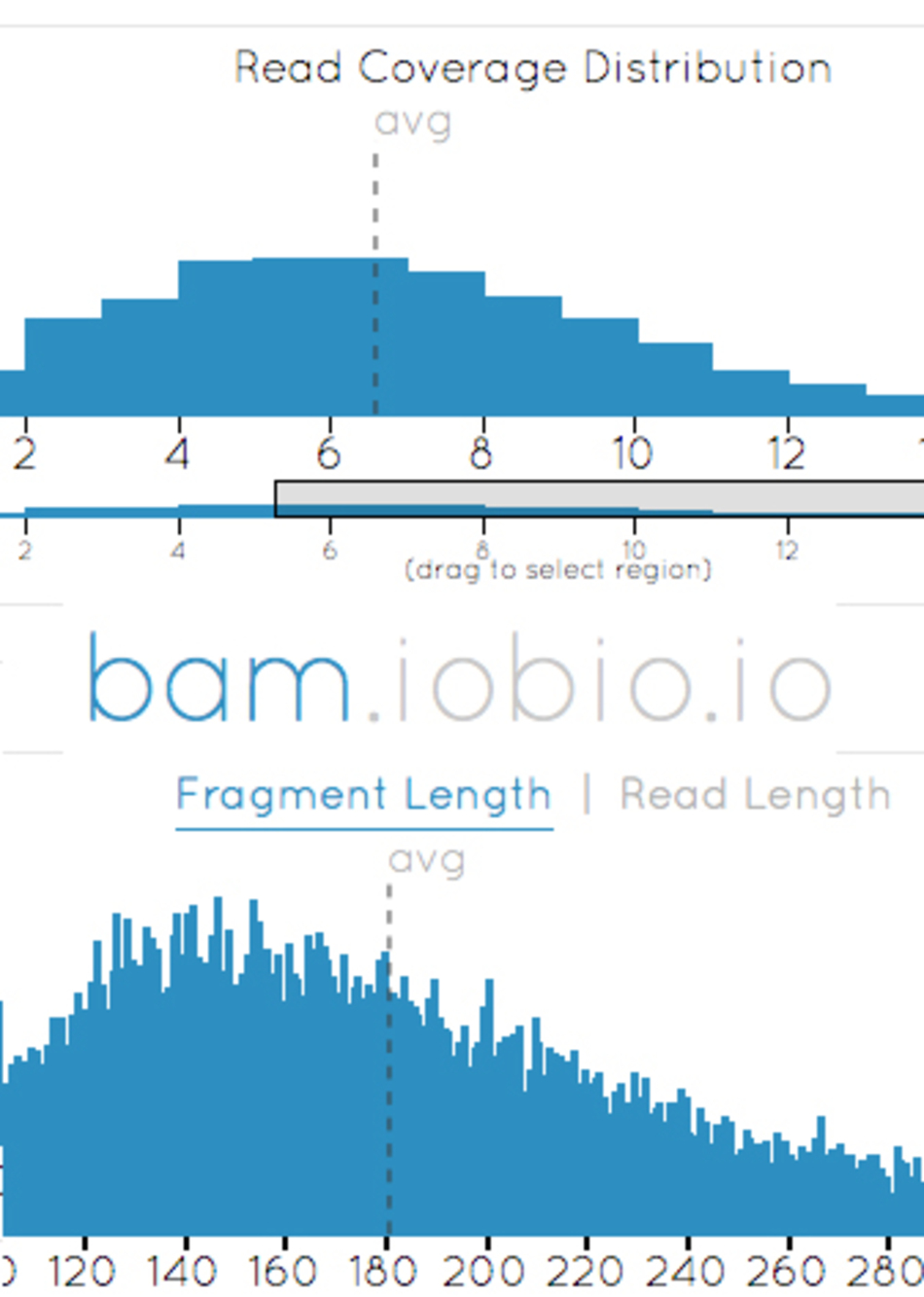
Dr. Gabor Marth: In a Bam file, you're communicating sequence alignments. That's the data that's generated by the sequencing machines and is the common currency of every type of genomic analysis.
This specific app enables the researcher to look at a sequence alignment file and within just a few seconds be able to look at many interesting aspects, for example how much data you have from a particular individual, what is the quality of your data. It enables you to go into detail into individual regions, look at individual chromosomes, and understand the overall quality of your data that all your downstream analysis will be based on.
Interviewer: So, this is kind of a first step. When you get your genomic sequence data back, you're checking it over to make sure that it's the quality you need and the coverage that you need.
Dr. Gabor Marth: That's the very first step. This is something that every researcher should do before they start analyzing the data any further.
Interviewer: How does this compare to what was available before?
Dr. Gabor Marth: Instead of doing genomic analysis end to end that would normally take days, weeks, and sometimes over a month, and require lots of computational resources and data storage and computer clusters, etcetera, the user just uses a laptop computer browser and is able to look at that part of the data that's important to them within a few seconds. Then further explore, go back and forth, change analysis parameters, etcetera, and redo analysis in real time as many times as they want and develop a real connection to the data that they analyze and the tools they use.
For example, sequence coverage, super important for a researcher is how much data do I have. We can estimate that in seconds. Using traditional UNIX-based tools, it would take hours to get the same answer.
Interviewer: So, it's kind of like going from one of the first Apple computers to an iPhone 6.
Dr. Gabor Marth: Well, that's a very flattering analogy but not entirely incorrect.
Interviewer: You know, if you do that comparison, that sort of iPhone 6 interactivity has been around for a while now, five years or so, why do you think it's taken so long for this type of analysis to catch up?
Dr. Gabor Marth: Genome scale, the ability to generate data on the scale of genome at ever decreasing costs, very cheaply today, has taken the community by surprise. The developer community focused on being able to analyze the data in its entirety and develop algorithms that are able to deal with the data on the scale that is produced today. That was the first impetus.
Now that we want to make this data count for the researchers is the time to start thinking about making it useful and making it useful without the type of computational investments, hardware investments that is very expensive and can only be done at institutional scale.
Interviewer: Who is this app designed for?
Dr. Gabor Marth: Bioinformaticians, core facilities that deliver data to other researchers, and researchers themselves. You pay for data. You get your data back from your core facility. The first thing you can do is check out the data quality.
Interviewer: These are part of a larger plan that you have.
Dr. Gabor Marth: In fact, we just completed a second app very similar in nature that allows you to look at genetic variants. This app is called vcf.iobio.io. It looks at VCF files. These are the files that programs create with genetic variants that they find, for example, in an individual human genome. This app, very similarly to bam.iobio, gives you overall genome-wide metrics, chromosome-wide metrics, or regional metrics of your file, things that are important to assess whether your variants are properly called and give you some regional understanding of your variants as well.
Our plan is to develop a very large number, an entire ecosystem, of genomic analysis apps that operate on the same exact principle. They will be able to help researchers with these more general data analysis tasks as well as more domain specific scientific tasks.
Interviewer: You're encouraging developers to tinker with Iobio.
Dr. Gabor Marth: Yes. What we are focusing on now that this paper is out is to develop the tools that will enable other developers to build genomic analysis apps of their own. Currently, to develop a genomic analysis app, it takes a long time, because you have to write pretty much everything from scratch. We'll be making libraries available that a developer can use to use what we call the Iobio platform. That is the type of software libraries that a developer needs to put together a prototype app maybe in a day and a more polished app in a couple of weeks.
We are still working on the developer support, so stay tuned. It will be two to three months before we are able to release the first versions of this.
Announcer: Interesting, informative, and all in the name of better health. This is The Scope Health Sciences Radio.