Episode Transcript
Interviewer: A new tool that could completely change how we diagnose infectious diseases, up next on The Scope.
Announcer: Examining the latest research and telling you about the latest breakthroughs, the Science and Research Show is on The Scope.
Interviewer: I'm talking with Gabor Marth, professor of human genetics at the University of Utah and co-director of the USTAR Center for Genetic Discovery.
Diagnosing infectious diseases is still a significant problem in medicine. Right now in the clinic, you either do a culture or maybe a PCR test to see what's infecting a person. You have co-developed a tool called Taxonomer that's taking a completely different approach.
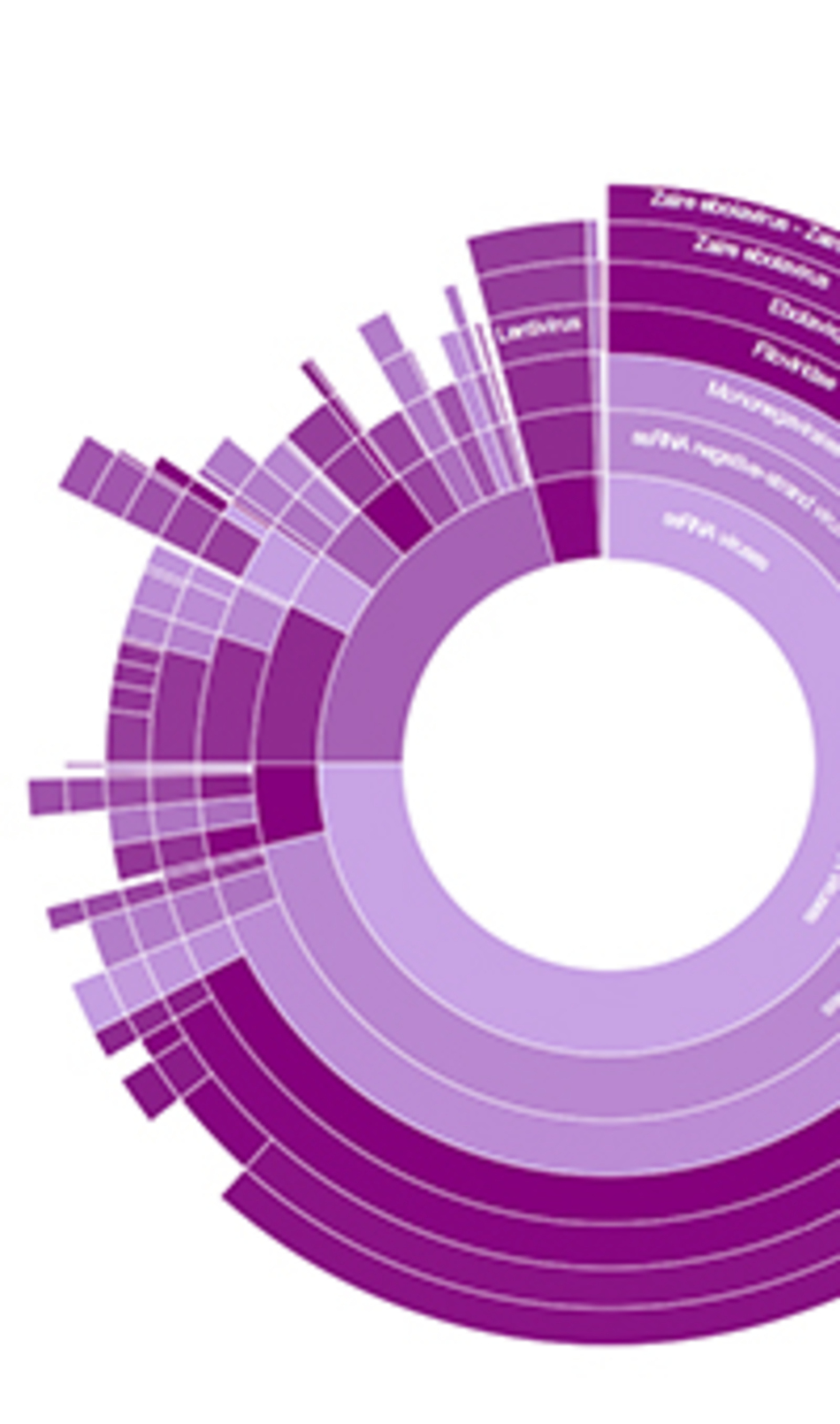
Gabor: Taxonomer is a software tool that is able to look at DNA fragments, which is what current technologies are able to produce from blood from an infectious disease patient, and identify the organisms that are present in that sample.
These types of tools, in my opinion, are very likely to replace the tests that you mentioned, that are based on specific hypotheses. We have to know what the strains that we are looking for in these patients, whereas tools like Taxonomer will be able to look at the DNA sequences and tell you upfront what pathogenic organisms are causing the patients disease.
Interviewer: So if you have a patient that's sick, you take a little blood, you have to get the DNA from that blood sequence for all the genetic material, I guess. And then that sequence information goes in the Taxonomer, and Taxonomer catalogues everything that's in there.
Gabor: Yes, as you mentioned, in traditional clinical tests you would have to ask "Does this patient have this particular strain of, which specific pneumonia strain this particular patient has." Sometimes these experiments fail. Some of the strains go better in the media that are used in laboratory tests than others. When the same data goes to a Taxonomer you just get an unbiased view of which strains and what proportions are present in the patient. That will help the physician diagnose the patient much faster, and in a much more objective manner.
Interviewer: So there are some really stand-out characteristics of Taxonomer. These are based on the software called Iobio, which is what you developed, correct?
Gabor: Taxonomer itself is the engine, it is the software that is able to take a DNA fragment and tell you which organism that DNA fragment presents. And it's married to Iobio, which is a web-based analysis platform that my laboratory is developing, that allows interactive, real-time and high visual representation of the data.
Interviewer: So yes, one way I've heard it described is that it's like putting a super computer in the hands of your average user, it has that much power behind it. But it's accessible on the internet.
Gabor: This is actually an accurate description because the computational power that allows Taxonomer to run is enormous but all that is hidden from the user. It's what's called the backhand of the architecture, there is a powerful computer sitting in the background that performs the computation in very, very intensive classification job. But what the users see is almost immediate response and almost immediate results.
Interviewer: And so before, this type of analysis took a lot longer and probably had to be done by a specialist, right? A bio-informatic specialist. But now anybody could do it. Is that the idea?
Gabor: Many, many things that the average user will be able to do. Of course, this is not to say that there is no longer need to for a . . .
Interviewer: We don't need you anymore.
Gabor: More involved analysis and expert analysis, but what Taxonomer will be able to do, it will be able to give a scientist, or even a lay-person, an immediate view of what is the overwhelming characteristics of the data. For example, we have examples of an Ebola patient. And when you look at the viral composition of the blood taken from this Ebola patient, you will see that pretty much 100% of the viral load in this individual is Ebola. So that's a very, very obvious and easily interpretable answer to the question of what's making this person sick.
Interviewer: Well like you said, a lot of the strength of this tool is that it's very visual, and that helps people take in information in a different way. Can you talk about that a little bit? Why you think that's an important component.
Gabor: That's how the human mind works. Experts can be trained to look at large data files and textual outputs coming from software and interpret that data in a scientifically meaningful fashion. But our human brain doesn't quite work that way. We like to see things, we like to use the brains innate analytical abilities to start from an image to develop an understanding. And that is the underlying philosophy behind the Iobio project.
Interviewer: In a few years how do you envision Taxonomer, in particular, being used?
Gabor: I view this that its applicability is going to be very wide. This is going to be a very wide and generally applicable tool. Some of the applications that we can foresee today will be infectious disease identification in the clinic that will require expert review of the data. Moving forward, the obvious application will be pathogen identification in the field, under field conditions.
Once the technologies are developed for portable and faster DNA sequence can be used under field conditions. This is only scratching the surface, checking sanitary conditions in restaurants and many applications that we can think of now and many even more applications that will become clear in the future.
Announcer: Interesting, informative, and all in the name of better health. This is The Scope Health Sciences Radio.